The report (and the data) from our Sloan-funded taxonomic annotation workshop on fungi in the built environment was just published in MycoKeys: https://mycokeys.pensoft.net/article/20887/list/4/
Abstract
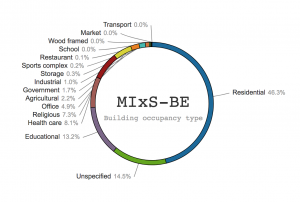
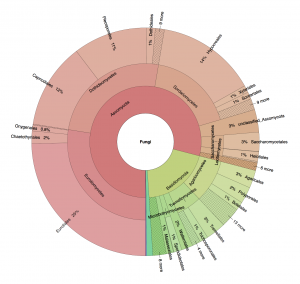
Recent DNA-based studies have shown that the built environment is surprisingly rich in fungi. These indoor fungi – whether transient visitors or more persistent residents – may hold clues to the rising levels of human allergies and other medical and building-related health problems observed globally. The taxonomic identity of these fungi is crucial in such pursuits. Molecular identification of the built mycobiome is no trivial undertaking, however, given the large number of unidentified, misidentified, and technically compromised fungal sequences in public sequence databases. In addition, the sequence metadata required to make informed taxonomic decisions – such as country and host/substrate of collection – are often lacking even from reference and ex-type sequences. Here we report on a taxonomic annotation workshop (April 10–11, 2017) organized at the James Hutton Institute/University of Aberdeen (UK) to facilitate reproducible studies of the built mycobiome. The 32 participants went through public fungal ITS barcode sequences related to the built mycobiome for taxonomic and nomenclatural correctness, technical quality, and metadata availability. A total of 19,508 changes – including 4,783 name changes, 14,121 metadata annotations, and the removal of 99 technically compromised sequences – were implemented in the UNITE database for molecular identification of fungi (https://unite.ut.ee/) and shared with a range of other databases and downstream resources. Among the genera that saw the largest number of changes were Penicillium, Talaromyces, Cladosporium, Acremonium, and Alternaria, all of them of significant importance in both culture-based and culture-independent surveys of the built environment.
Figures and figure legends
Many thanks to all workshop participants for making this happen!
