A while back we posted about our microBEnet project to have undergraduates come into the lab and sequence reference genomes from the built environment. That project now has it’s own blog, being maintained by the students themselves. Comments about our original post led to the following guest post by Paul Orwin, who is doing something very similar at CSUSB.
Guest post by Paul Orwin, Associate Professor of Biology, CSU San Bernardino.

The California State University at San Bernardino is a regional comprehensive university in the Inland Empire region of California (Riverside and San Bernardino Counties). It is one of only two (the other being UC Riverside) public universities serving this region. As a master’s level school, we have a diverse student body from a lot of different educational and ethnic backgrounds. Designing courses for this group of students is a challenge! Fortunately, by the time they reach the upper division Biology courses, they have had a thorough grounding in biological sciences and chemistry. This makes my task in putting together this course easier. Many of the students see themselves following a health professions route in the future, including Medical, Dental, and Pharmacy school as well as some interested in Ph.D. studies. So there is a lot of demand for a course in Medical Microbiology, but I wanted to spice things up a bit! I decided that where I could do that was in the laboratory segment of the class, by including an enrichment and isolation experiment along with the traditional clinical microbiology diagnostic experiments.
I first need to explain where the idea for this course came from. For several years I have been taking students from my lab group to the International Conference on Microbial Genomics held at Lake Arrowhead, CA every other year. This is a fantastic meeting, organized by Jeffrey H. Miller (gotta be careful with those middle initials!) at UCLA. In a not terribly surprising coincidence, Jonathan Eisen (who got me interested in microBEnet) and Ashlee Earl (who I don’t think has a web site) (who will appear later in this story) are involved in organizing it this year. At this meeting I learned a great deal about genomics and metagenomics, and got interested in the idea of incorporating this type of work into the classroom based on the work Jeffrey Miller and Erin Sanders were doing with UCLA Microbiology undergraduates. One year they reported on their efforts to sequence and annotate the genome of a novel microorganism and another time Erin’s class put up posters describing the phage they identified, sequenced, and annotated. They wrote a textbook about this work, which goes to show they are dedicated to this idea! As we will see, I have not gotten nearly that far in my own efforts. Another source of inspiration for this work was the class that Jared Leadbetter taught at CalTech when I was working with him, on enrichment and isolation strategies from the environment (including the Built Environment, incidentally). The inventiveness of these students was remarkable, as was the frequency with which they were successful. Of course, he has forgotten more microbiology than I will ever know, which probably helps. After I started my own faculty journey, I drew on this inspiration as well as many conversations in various forms with Mark Martin (a true microbial supremacist) to develop an enrichment and isolation approach for my general Microbiology course. Mark and Jared (and others) have inspired me to think about culture techniques, and about the claim that much of the microbiome is “unculturable” (preposterous, IMHO).
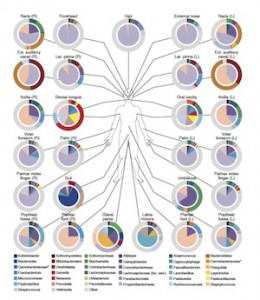
The final person who got me interested in this is the aforementioned Dr. Ashlee Earl, who presented some work on the Human Microbiome Project at the last ASM general meeting. On her poster, she described how the HMP had identified a group of 100 most wanted organisms — organisms that they wanted other labs and research groups to isolate so that a good set of reference genomes could be developed. This served as the jumping off point for my course design (if you can call it that).
Ok, the name dropping is out of the way (or the giving credit where credit is due, if you prefer), so on to the class itself. It is a class in Medical Microbiology, with the lecture based on Mims’ Medical Microbiology. The lab is based on enrichment and isolation techniques, bringing together classical clinical microbiology tests (metabolic testing, serotyping, and staining) with 16S rRNA sequence analysis for identification purposes. The idea here is to teach the students how to use these techniques for two major things a medical microbiologist might do — identify a known pathogen by rapid testing procedures, or identify and classify an unknown organism associated with a pathology.
The first half of the course (which we have just completed) involved identifying organisms from a mixed culture (given to the students by myself) based on traditional microbiological techniques. This identification was complemented with a 16s rRNA experiment, which also served as an introduction/refresher on basic molecular biology techniques (PCR, gel electrophoresis, DNA extraction). When the DNA sequences are returned to us from the sequencing facility, we will be analyzing them using the RDP database. This will also give us a chance to discuss error in sequencing and PCR, as well as the difference between identifying and classifying. Hopefully they get the same thing from the sequencing as they got from the culture tests!
We have spent a good deal of time discussing the idea of enrichment and isolation, and how this can be applied to the Human Microbiome. They have seen the immense diversity of the microbiome (cite) as well as the difference between what is there and what is published. To prepare them for the task, I used the HMP table that lists off the organisms identified from various body sites and categorizes them as “Most Wanted, Medium Priority, and Low Priority.” I just gave the students the “Most Wanted” organisms to work with, and to make things a little more comfortable for them I eliminated the stool sample organisms. I then proposed several options to them.
1) Everyone could agree on a single target to isolate, and we could design a number of different media to try to enrich for and isolate these bacteria.
2) Everyone could go their own way, picking individual organisms and designing experiments to enrich, isolate, and identify them.

In the end, several students chose to go their own way, while a number chose to focus on one group (the oral actinomycetes) and come up with multiple different approaches to isolate these bacteria. They all did background research on what is known about culturing these organisms from various sources, and we all agreed on using three of complementary strategies to enrich and isolate these bacteria. The first approach is the traditional enrichment, based on known characteristics of the species, design media that encourage actinomycetes (like potato agar) to grow. The second is to use the desiccation tolerance of the actinomycete spore as a strong selection against other vegetative cells (this is riskier, since there are endospore formers present as well, and we don’t know if the actinomycetes in the oral microbiota sporulate. The final approach is my personal favorite — the oligotrophy approach. First, put them on media with nothing in it (perhaps trace minerals). Let micro colonies form on that plate, then pick the microcolonies onto separate “nothing medium” and let them grow in isolation. Finally carefully pick them onto rich medium (or maybe just a bit richer medium, like 100 mg/L YE) to let them grow big enough to test. If they grow well on rich medium, we can do biochemical tests, or we can just go with the molecular identification. I can’t take credit for this idea (I first heard it from Jared), but I’ve used it a few times and I like it. It helps find bacteria that don’t grow very fast on rich media, or get outcompeted by the boring old familiars on typical clinical microbiology medium.
So that’s where we are right now, with IRB approval in hand, ready to embark on the adventure. I think it will work, and I’m sure that we will all learn something! Sadly I am not enough of a tech geek (yet!) to have the students blogging or tweeting the experience. Maybe next year…
Very cool! How are you dealing with the biosafety issues involved – is this all in BSL-2?
Apologize for the tardiness of the response – I just noticed the comment. Yes, we treat everything as BSL-2. Based on what I know about this type of expt, that should be sufficient. We are trying to avoid the bad guys – most of the students are using an actinomycete enrichment strategy, which should be pretty safe, I think.