Old photographs often are unique artifacts that provide a realistic look into the recent human history. However, many of these older printed photos are deteriorating, due to humidity, temperature, extended light exposure, and biodeterioration. Scientific Reports published a nice paper this week about the microbes found on and in old photographs, called Microbial communities affecting albumen photography heritage: a methodological survey by Puškárová et al.
In this study, the authors (from Slovakia) first investigated the chemical composition of two photographs, both dating from around the year 1900. Both photographs contained albumen, a protein found in egg whites, which was used around that period to bind the light-sensitive halide salts to a paper underlayer. It’s easy to imagine that albumen pictures are supportive of microbial growth. Indeed, both photographs showed “foxing” spots, brownish spots often found on old paper documents or painting.
Microscopic analysis revealed that the foxing spots in both photographs contained evidence of fungal colonization, i.e., hyphae and spores.
Microbial communities were analyzed by culturing membranes pressed against certain areas of the pictures and the surrounding paper of photograph #1, and subsequent amplification with broad-range bacterial or fungal primers. In addition, the membranes were analyzed by direct PCR of their extracted DNA, followed by DGGE and sequencing of clone libraries.
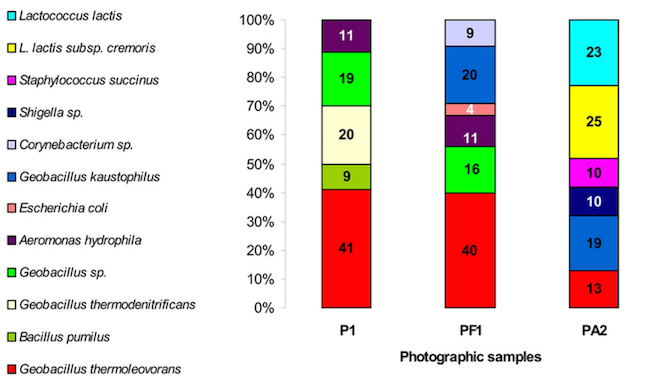
The bacterial communities from both photographs contained Geobacillus spp, but were otherwise very different bacterial communities; Photo 1 contained mainly Geobacillus spp., Aeromonas hydrophila and Bacillus pumilus, while Photo 2 yielded Lactococcus lactus, Geobacillus, Staphylococcus and Shigella spp..
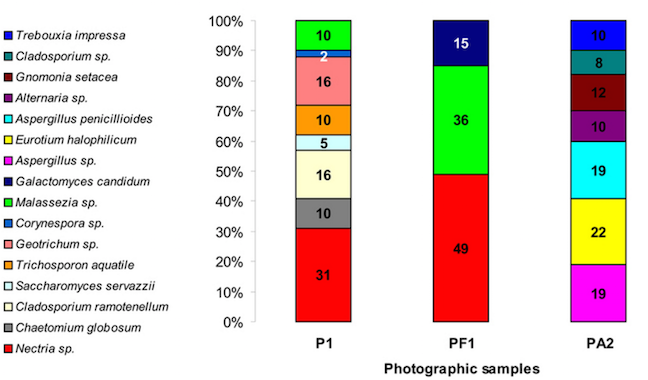
Similarly, the fungal communities of the two photographs showed differences too. Both Photo 1 as well as the surrounding paper contained Nectria and Malassezia, but the albumen fungal diversity was much greater. Photo 2’s fungal communities consisted of Aspergillus, Eurotium, Gnomonia, Alternaria, and Cladosporium.
The study also demonstrates biodegradation activities in the isolated bacteria and fungi. For example, the Bacillus from Photo 1 showed intense lipase and cellulase activity. The fungi isolated from both photos were less aggressive.
The authors conclude that analyses like these could help in better understanding and eventually preventing the deterioration of albumen prints.
Safeguarding our cultural heritage requires methods able to identify the construction materials of historical objects and to diagnose the potential biodeteriogens in order to develop targeted conservation and restoration strategies.
 A similar analysis of “foxing” spots was done on a self-portrait of Leonardo Da Vinci: Amid the possible causes of a very famous foxing: molecular and microscopic insight into Leonardo da Vinci’self-portrait by Piñar et al.; I wrote about that paper in an August 2015 MicroBEnet post. The foxing spots in that paper were mainly composed of Eurotium halophilicum, Ascomycota, and Acremonium spp.
A similar analysis of “foxing” spots was done on a self-portrait of Leonardo Da Vinci: Amid the possible causes of a very famous foxing: molecular and microscopic insight into Leonardo da Vinci’self-portrait by Piñar et al.; I wrote about that paper in an August 2015 MicroBEnet post. The foxing spots in that paper were mainly composed of Eurotium halophilicum, Ascomycota, and Acremonium spp.
Microbial communities affecting albumen photography heritage: a methodological survey
Andrea PuÅ¡kárová, Mária BuÄková, Božena Habalová, Lucia Kraková, Alena Maková & Domenico Pangallo – Scientific Reports
Full text of the abstract:
This study is one of the few investigations which analyze albumen prints, perhaps the most important photographic heritage of the late 19th and early 20th centuries. The chemical composition of photographic samples was assessed using Fourier-transform infrared spectroscopy and X-ray fluorescence. These two non-invasive techniques revealed the complex nature of albumen prints, which are composed of a mixture of proteins, cellulose and salts. Microbial sampling was performed using cellulose nitrate membranes which also permitted the trapped microflora to be observed with a scanning electron microscope. Microbial analysis was performed using the combination of culture-dependent (cultivation in different media, including one 3% NaCl) and culture-independent (bacterial and fungal cloning and sequencing) approaches. The isolated microorganisms were screened for their lipolytic, proteolytic, cellulolytic, catalase and peroxidase activities. The combination of the culture-dependent and -independent techniques together with enzymatic assays revealed a substantial microbial diversity with several deteriogen microorganisms from the genera Bacillus, Kocuria, Streptomyces and Geobacillus and the fungal strains Acrostalagmus luteoalbus, Bjerkandera adusta, Pleurotus pulmonarius and Trichothecium roseum.