I made a Storify summary of the Tweets from a workshop at the Pacific Symposium on Biocomputing that I attended on Tuesday on Computational Microbiology (with a big focus on microbiomes). [<a href="//storify.com/phylogenomics/psb16-session-on-microbiomes” target=”_blank”>View the story “#PSB16 session on computational microbiome #microbiomes ” on Storify]
Here are my notes from day 3 of the Mothur Workshop that is taught by Pat Schloss (pdschloss at gmail.com) at a hotel that is conveniently located near the Detroit airport. Of all of the bioinformatic workshops I have attended, this group of students was my favorite so far. We had a lot of fun …
Here are my notes from day 2 of the Mothur workshop taught by Pat Schloss (pdschloss at gmail.com) in December 2015. For those who are interested in learning to use Mothur for microbiome studies, Pat will be teaching another one in February. Mothur is better for bacterial characterization than eukaryotes because the sequences are aligned before OTU clustering. This …
Pat Schloss (pdschloss at gmail.com) offers excellent workshops on the Mothur software for analyzing 16S rRNA data for bacteria. He’s just announced the next one (February 8 to 10, 2016 near the Detroit airport). I had the pleasure of attending the December workshop. A diverse and international group attended the workshop, with many folks who are interested in the human microbiome. …
Blog post prepared jointly by Andrew Doxey (@acdoxey) and Josh Neufeld (@joshdneufeld) The “aquariome” Back in 2013, as part of a project assessing aquarium microbial communities and their role in nutrient cycling, Laura Sauder (graduate student in the Neufeld lab) sequenced a shotgun metagenomic library from a freshwater aquarium biofilter that was installed on this …
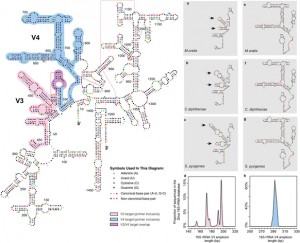
To date, characterization of ancient oral (dental calculus) and gut (coprolite) microbiota has been primarily accomplished through a metataxonomic approach involving targeted amplification of one or more variable regions in the 16S rRNA gene. For anyone interested in rRNA studies of microbial communities, ancient microbiomes, or analysis of samples with small amounts of DNA this …
Gearing up the UNITE database for the built mycobiome The team behind the UNITE database for molecular identification of fungi has been granted support from the Sloan Foundation to strengthen the support for fungi from the built environment. Launched in 2001 as an ITS database for identification of ectomycorrhizal fungi in the Nordic countries, UNITE …
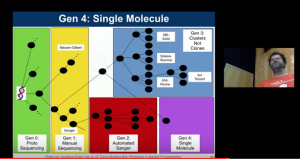
Every year for the last few years I have given a talk on the “Evolution of DNA Sequencing” at the “Workshop in Applied Phylogenetics” at Bodega Bay Marine Lab. I just did the talk and thought I would post the slides here. I note – I also added an evolutionary tree of sequencing methods which …
Norm Pace gave a talk at UC Davis yesterday on “Metagenomics and the Tree of Life”. I and a few other people posted live Tweets from the talk which I have compiled together via the Storify system. This “Storify” is embedded below. In addition, Lisa Cohen, who was at the talk posted her notes which …
scikit-bio is a library implementing core bioinformatics algorithms and data structures, and providing support for bioinformatics data munging. You can learn more about the project by watching Jai Rideout and Evan Bolyen’s 20 minute SciPy 2015 talk. scikit-bio currently supports Python 2 (i.e., Legacy Python) and Python 3. We’re considering dropping support for Python 2, …