Public fungal sequence data are often not annotated to the degree desirable. This makes it difficult to zoom in on all sequences recovered from some particular country or some specific host. This holds true for the built environment as well – there is no straightforward way to target all fungal sequences generated from, say, bathrooms or gypsum board.
To mitigate these concerns, a sequence annotation workshop was organized at the University of Gothenburg in May 2016. The 36 workshop participants sought to annotate all public fungal barcode (ITS) sequences from the built environment according to the MIxS-BE standard and to make the annotations public through the UNITE database and other online resources.
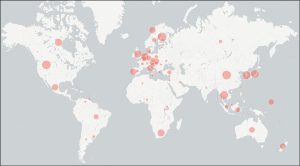
A report from the workshop was just published in MycoKeys. A total of 45,488 data points were assembled from the literature, including 8,430 instances of country of collection from a total of 83 countries (Fig. 1), 5,801 instances of building types (Fig. 2), and 3,876 instances of surface-air contaminants. The workshop participants managed to assemble at least one data point for nearly all public Sanger-derived fungal ITS sequences from the built environment. The participants were sometimes surprised by how hard it was to pinpoint the targeted data in the scientific publications – a reminder to us all to write our scientific reports in a clear, standardized way.
The workshop participants also ran into situations and built environments that were somewhat difficult to represent in the MIxS-BE standard in a detailed way – the examples include spacecraft, mummy crypts, and pigeon houses. The participants furthermore felt that some aspects of the sampling of indoor air and floors were not easy to squeeze into the standard. A list with 14 potential amendments to the MIxS-BE standard was composed and forwarded to the MIxS-BE consortium.
The data assembled during the workshop were uploaded into the PlutoF workbench of UNITE to support detailed queries of the built mycobiome. The data were furthermore shared with a range of online resources and are bundled with the publication. The workshop was funded through a Sloan grant to the UNITE team and was organized jointly by the UNITE and ISHAM databases.
Figure 1. Analysis of the built mycobiome sequences for country of collection.
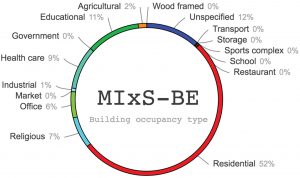 Figure 2. Analysis of the MIxS-BE “building occupancy type” (type of building where the underlying sample was taken).
Figure 2. Analysis of the MIxS-BE “building occupancy type” (type of building where the underlying sample was taken).